Systems Genomics - Heinäniemi lab
Cells represent the fundamental unit of life. Within tissues, cells are in constant interaction with their environment. Our goal is to elucidate disease mechanisms that are related to aberrant regulation of cell states. We develop computational models based on systems biology and data analysis tools with bioinformatics and machine learning methods suitable for integrating different types of genomics data or combining datasets across studies. We apply them in our own research that utilizes large omics datasets and collect new genomics data from cell models and human studies.
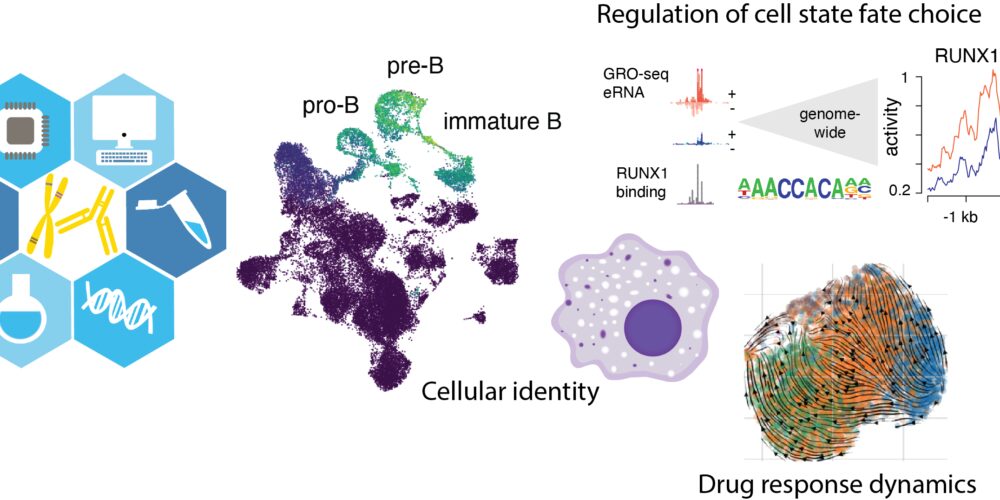
Childhood acute lymphoblastic leukemia: The most common type of cancer in children develops as a consequence of arrested cell differentiation of early lymphoid precursor cells. We study changes in the gene regulatory network across different leukemia subtypes. To translate genomics into clinical practice, we also develop data analysis tools that allow determining at unprecedented resolution the alterations in cancer cells.
Cell-cell interaction in disease development: In addition to regulatory networks that operate inside cells, we study how these interactions extend to tissue level. Inflammation and cellular stress influence cell states within a tissue. We are addressing the question what role changes in cell-cell-interactions have in this process. By characterizing such tissue-level regulatory circuits, we can discover new therapy targets that would enable reverting the changes in cell states at early stage during disease progression. Later at advanced disease state it would be more challenging to repair tissue damage. We are focusing on the role of endothelial cells and macrophages in context of coronary artery disease development. Cell-cell interactions have an important role also in cancer.
A central future goal in our research is to connect the molecular and cell-interaction levels together, in order to better model the dynamics of cell state changes.
Publications – selected publications; all publications co-authored by principal investigator in orcid
News
-
 Merja Heinäniemi appointed as Professor of Computational Biomedicine
Merja Heinäniemi appointed as Professor of Computational BiomedicineMerja Heinäniemi appointed as Professor of Computational Biomedicine
-
 Better treatments for leukemia
Better treatments for leukemiaBetter treatments for leukemia
Keywords
Professors
Post-doctoral Researchers
Doctoral Researchers
-

Mari Lahnalampi
Postdoctoral ResearcherInstitute of Biomedicine, School of Medicine, Faculty of Health Sciences -
Sanni Moisio
Doctoral ResearcherInstitute of Biomedicine, School of Medicine, Faculty of Health Sciences -
Andrea Hanel
Doctoral ResearcherInstitute of Biomedicine, School of Medicine, Faculty of Health Sciences -
Buddika Jayasingha
Doctoral ResearcherInstitute of Biomedicine, School of Medicine, Faculty of Health Sciences -

Konstantin Ivanov
Doctoral ResearcherInstitute of Biomedicine, School of Medicine, Faculty of Health Sciences
Project researcher
Publications
9 items-
In vivo interrogation of regulatory genomes reveals extensive quasi-insufficiency in cancer evolution
Fischer, Anja; Lersch, Robert; de Andrade Krätzig, Niklas; Strong, Alexander; Friedrich, Mathias J.; Weber, Julia; Engleitner, Thomas; Öllinger, Rupert; Yen, Hsi-Yu; Kohlhofer, Ursula; Gonzalez-Menendez, Irene; Sailer, David; Kogan, Liz; Lahnalampi, Mari; Laukkanen, Saara; Kaltenbacher, Thorsten; Klement, Christine; Rezaei, Majdaddin; Ammon, Tim; Montero, Juan J.; Schneider, Günter; Mayerle, Julia; Heikenwälder, Mathias; Schmidt-Supprian, Marc; Quintanilla-Martinez, Leticia; Steiger, Katja; Liu,. 2023. Cell genomics. 3: A1 Journal article (refereed), original research -
Combination therapies to inhibit LCK tyrosine kinase and mTOR signaling in T-cell Acute Lymphoblastic Leukemia
Laukkanen, Saara; Bacquelaine Veloso, Alexandra; Yan, Chuan; Oksa, Laura; Alpert, Eric J.; Do, Daniel; Hyvärinen, Noora; McCarthy, Karin; Adhikari, Abhinav; Yang, Qiqi; Iyer, Sowmya; Garcia, Sara P.; Pello, Annukka; Ruokoranta, Tanja; Moisio, Sanni; Adhikari, Sadiksha; Yoder, Jeffrey A.; Gallagher, Kayleigh Mary; Whelton, Lauren; Allen, James R.; Jin, Alexander Hai; Loontiens, Siebe; Heinäniemi, Merja; Kelliher, Michelle A; Heckman, Caroline A.; Lohi, Olli; Langenau, David M. 2022. Blood. 140: 1891–1906 A1 Journal article (refereed), original research -
Genome-wide analysis of primary microRNA expression using H3K36me3 ChIP-seq data
Turunen, Tanja; Hernández de Sande, Ana; Pölönen, Petri; Heinäniemi, Merja. 2021. Computational and Structural Biotechnology Journal. 19: 1944-1955 A1 Journal article (refereed), original research -
Germline POT1 Deregulation Can Predispose to Myeloid Malignancies in Childhood
Michler, Pia; Schedel, Anne; Witschas, Martha; Friedrich, Ulrike Anne; Wagener, Rabea; Mehtonen, Juha; Brozou, Triantafyllia; Menzel, Maria; Walter, Carolin; Nabi, Dalileh; Pearce, Glen; Erlacher, Miriam; Göhring, Gudrun; Dugas, Martin; Heinäniemi, Merja; Borkhardt, Arndt; Stölzel, Friedrich; Hauer, Julia; Auer, Franziska. 2021. International journal of molecular sciences. 22: 11572 A1 Journal article (refereed), original research -
Immunogenomic Landscape of Hematological Malignancies
Dufva, Olli; Pölönen, Petri; Brück, Oscar; Keränen, Mikko AI; Klievink, Jay; Mehtonen, Juha; Huuhtanen, Jani; Kumar, Ashwini; Malani, Disha; Siitonen, Sanna; Kankainen, Matti; Ghimire, Bishwa; Lahtela, Jenni; Mattila, Pirkko; Vähä-Koskela, Markus; Wennerberg, Krister; Granberg, Kirsi; Leivonen, Suvi-Katri; Meriranta, Leo; Heckman, Caroline; et al. [incl. Heinäniemi, Merja]. 2020. Cancer cell. 38: 380-399.e13 A1 Journal article (refereed), original research -
Single cell characterization of B-lymphoid differentiation and leukemic cell states during chemotherapy in ETV6-RUNX1-positive pediatric leukemia identifies drug-targetable transcription factor activities
Mehtonen, Juha; Teppo, Susanna; Lahnalampi, Mari; Kokko, Aleksi; Kaukonen, Riina; Oksa, Laura; Bouvy-Liivrand, Maria; Malyukova, Alena; Mäkinen, Artturi; Laukkanen, Saara; Mäkinen, Petri I; Rounioja, Samuli; Ruusuvuori, Pekka; Sangfelt, Olle; Lund, Riikka; Lönnberg, Tapio; Lohi, Olli; Heinäniemi, Merja. 2020. Genome medicine. 12: Article number: 99 A1 Journal article (refereed), original research -
Hemap: An interactive online resource for characterizing molecular phenotypes across hematologic malignancies
Pölönen, P; Mehtonen, J; Lin, J; Liuksiala, T; Häyrynen, S; Teppo, S; Mäkinen, A; Kumar, A; Malani, D; Pohjolainen, V; Porkka, K; Heckman, CA; May, P; Hautamäki, V; Granberg, KJ; Lohi, O; Nykter, M; Heinäniemi, M. 2019. Cancer research. 79: 2466-2479 A1 Journal article (refereed), original research -
Semisupervised Generative Autoencoder for Single-Cell Data
Ngo Trong, Trung; Mehtonen, Juha; González, Geraldo; Kramer, Roger; Hautamäki, Ville; Heinäniemi, Merja. 2019. Journal of computational biology. [Published online: 2 December 2019]: 1-14 A1 Journal article (refereed), original research -
Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture
Bouvy-Liivrand Maria, Hernández de Sande Ana, Pölönen Petri, Mehtonen Juha, Vuorenmaa Tapio, Niskanen Henri, Sinkkonen Lasse, Unelma Kaikkonen Minna, Heinäniemi Merja. 2017. Nucleic acids research. 45: 9837-9849 A1 Journal article (refereed), original research