
Data-Driven Drug Design
Leaders
In the Data-Driven Drug Design (4D) group, we focus on bringing physics-based modeling strategies together with modern AI and data science approaches to accelerate the discovery of new therapeutics. Our work is inherently computationally intensive, and our research relies heavily on the use of the national and EuroHPC supercomputing resources to enable large-scale virtual screening, long-timescale molecular dynamics simulations, and a rapid iteration on different data-intensive workflows.
The group develops and applies methods and workflows for classical computer-aided drug discovery and molecular modeling, ultra-large-scale virtual screening, and efficient chemical space exploration. We are particularly passionate about data: We systematically curate, integrate and mine available experimental and computational knowledge to guide our decision-making, benchmark our tools, and fine-tune our workflows. This is complemented by our efforts in research software development and research into explainable AI strategies that provide intuitive, medicinal-chemistry-friendly insight into model decisions. Our approaches are applied across a range of anti-bacterial, anti-viral and anti-parasitic targets, as well as broader human health–relevant systems, and we collaborate closely with partners from academia and industry.
News
-
 Drug discovery on an unprecedented scale
Drug discovery on an unprecedented scaleDrug discovery on an unprecedented scale
Boosting virtual screening with machine learning allowed for a 10-fold time reduction in the processing of 1.56 billion drug-like molecules.… -
 4D at NoBSC 2026
4D at NoBSC 20264D at NoBSC 2026
Join 4Ders at the Nordic Basic Scientific Computing conference!
Cooperation
-
Heikki Käsnänen
Head of Molecular Prospecting and Modeling
-
-
Thales Kronenberger
Computational Drug Development group
-
 Molecular Modeling and Drug Design Research Group 01.01.2010 -
Molecular Modeling and Drug Design Research Group 01.01.2010 - -
 Asquith Medicinal Chemistry Group 01.01.2022 -
Asquith Medicinal Chemistry Group 01.01.2022 -
-
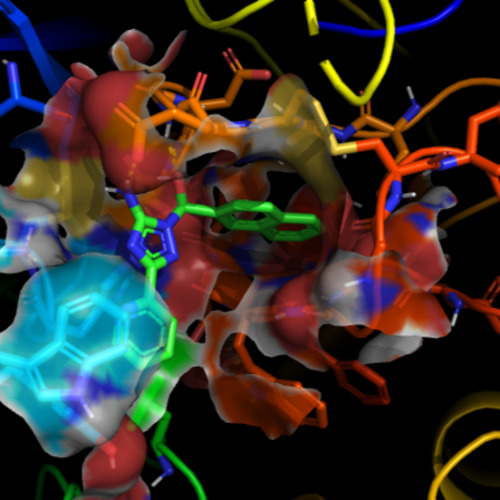
Proteins are dynamic and constantly fluctuate between many shapes. Ligand binding is influenced by these motions and often insufficiently modeled by a single static structure. Accounting for the target dynamics has become crucial in drug discovery: Both binding affinity and selectivity can depend on transient pockets, hidden conformations and subtle allosteric changes virtually invisible from single crystal structures alone. Our current projects explore ways to combine conventional approaches with AI-driven strategies to consider conformational ensembles and target dynamics even when running large-scale screening efforts.
FIKSU-AI
In the FIKSU-AI project, we explore flexible and ensemble docking with multiple protein conformations to study how ligands interact with different states of the target. Our aim is to accelerate existing physics‑based docking protocols with AI. This helps us to understand when learned models can be sufficient and when conventional docking concepts are still key to a successful virtual screening campaign for dynamic targets.
MAMMUTTI
In the MAMMUTTI project, we utilize long-timescale molecular dynamics simulations of multimeric proteins to sample their relevant rearrangements over time. From these trajectories, we aim to identify and quantify key conformational states of the target that matter for ligand binding and use these representative structures to guide docking-based virtual screening. In addition, we seek novel ways of using AI-driven strategies to support and speed up the screening process.
-
This section is under development. Tools and details are coming soon…
Also check our GitHub repository to explore more!
Keywords
Publications
6 items-
A Twist of Fate: The Helix–Turn–Helix Motif in Pseudomonas aeruginosa ExsA Can Allosterically Stabilize the Ligand-Binding Domain
Medarametla, Prasanthi; Greenhalgh, Jack Calum; Pöhner, Ina; Welch, Martin; Poso, Antti; Kronenberger, Thales; Rahman, Taufiq, 2025, Journal of chemical information and modeling, 65, 22, 12448-12463. A1 Journal article (refereed), original research -
Artificial intelligence in drug design: why a ‘one-size-fits-all’ approach remains out of reach
Almeida, Rafael Lopes; Campera, Gabriella Matos; Pöhner, Ina; Maltarollo, Vinicius Gonçalves, 2025, Expert opinion on drug discovery, 20, 10, 1239-1250. A1 Journal article (refereed), original research -
Ultra-Large-Scale Virtual Screening
Pöhner, Ina; Sivula, Toni; Poso, Antti, 2025, Maltarollo, Vinícius Gonçalves, Computer-Aided Drug Discovery and Design: From Theory to Applications, 299-343. A3 Book section, Chapters in research books -
Drug repurposing platform for deciphering the druggable SARS-CoV-2 interactome
Bogacheva, Mariia S; Kuivanen, Suvi; Potdar, Swapnil; Hassinen, Antti; Huuskonen, Sini; Pöhner, Ina; Luck, Tamara J; Turunen, Laura; Feodoroff, Michaela; Szirovicza, Leonora; Savijoki, Kirsi; Saarela, Jani; Tammela, Päivi; Paavolainen, Lassi; Poso, Antti; Varjosalo, Markku; Kallioniemi, Olli; Pietiäinen, Vilja; Vapalahti, Olli, 2024, Antiviral research, 223, 105813. A1 Journal article (refereed), original research -
The comprehensive SARS-CoV-2 ‘hijackome’ knowledge base
Huuskonen, Sini; Liu, Xiaonan; Pöhner, Ina; Redchuk, Taras; Salokas, Kari; Lundberg, Rickard; Maljanen, Sari; Belik, Milja; Reinholm, Arttu; Kolehmainen, Pekka; Tuhkala, Antti; Tripathi, Garima; Laine, Pia; Belanov, Sergei; Auvinen, Petri; Vartiainen, Maria; Keskitalo, Salla; Österlund, Pamela; Laine, Larissa; Poso, Antti; Julkunen, Ilkka; Kakkola, Laura; Varjosalo, Markku, 2024, Cell discovery, 10, 1, 125. A1 Journal article (refereed), original research -
Machine Learning-Boosted Docking Enables the Efficient Structure-Based Virtual Screening of Giga-Scale Enumerated Chemical Libraries
Sivula, Toni; Yetukuri, Laxman; Kalliokoski, Tuomo; Käsnänen, Heikki; Poso, Antti; Pöhner, Ina, 2023, Journal of chemical information and modeling, 63, 18, 5773-5783. A1 Journal article (refereed), original research

