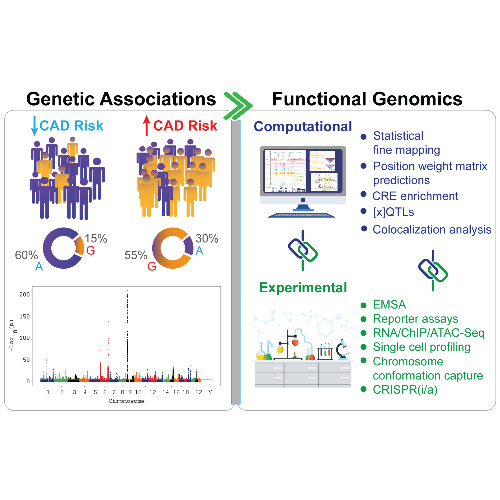
Our Cardiovascular Genomics lab investigates the mechanistic basis of Atherosclerotic Cardiovascular Disease (ASCVD) and cardiometabolic diseases, using a combination of cellular and genetic model systems, single-cell transcriptomics and epigenetics, high-throughput functional genomics techniques, and bioinformatics. We have unique expertise in functional fine-mapping of non-coding GWAS variants using massively parallel reporter assays and cellular models for CRISPR/Cas9-mediated perturbation. This has allowed us to perform the most extensive functional fine-mapping of ASCVD GWAS variants and provide unique insights into the genetic basis of disease. Our lab also hosts a single-cell genomics core and we are committed to leveraging the power of single-cell multiomics and spatial transcriptomics to drive cutting-edge research in the future.
Our research group operates in a highly interdisciplinary environment, where we bridge the fields of basic research in gene regulation and vascular biology, genetics, and bioinformatics. We have extensive collaboration networks with other world-leading labs, providing unique access to large clinic- and population-based genetic studies and cohorts of ASCVD and prospective cross-sectional patient biobanks. We are excited to continue pushing the boundaries of cardiovascular genomics research, and we invite you to join us on this journey!
News
Keywords
Professors
Senior Researchers
Post-doctoral Researchers
-

Uma Thanigai Arasu
Postdoctoral ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -

Mari Taipale
Senior ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -

Prosanta Singha
Project Researcher
Doctoral Researchers
-

Ilakya Selvarajan
Doctoral Researcher -

Aarthi Ravindran
Early Stage ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -
Eloi Schmauch
Grant-funded ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -
Anu Nyberg
Doctoral ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -
Valtteri Nurminen
Doctoral ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences -
Negar Pouyanfar
Doctoral ResearcherA.I. Virtanen Institute for Molecular Sciences, Faculty of Health Sciences
Technicians
Other group members
-
Iida Puranen ipuranen@student.uef.fi
-
Jimi Virkkala jimi.virkkala@uef.fi
-
Olayinka Oluwasegun Agbabiaje olayagba@student.uef.fi
-
Johannes Ojanen johannes.ojanen@aalto.fi



